Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
A Single-Lead Electrocardiogram-Derivative Empirical Mode Decomposition-Based Deep Learning Model for Sleep Apnea Identification
Authors: R. Krithiga Sree, Dr. D. Somasundareswari
DOI Link: https://doi.org/10.22214/ijraset.2024.63676
Certificate: View Certificate
Abstract
While polysomnography (PSG) is the gold standard for detecting sleep apnea (SA), the insertion of several disruptive devices may impair the quality of the patient\'s sleep, and its interpretation requires specialised training from a sleep scientist or technician. Heart rate variability (HRV) and electrocardiogram (ECG)-derived respiration (EDR) have been used in recent years to automatically detect SA and lessen the negative effects of PSG. Currently, the majority of suggested methods concentrate on feature engineering and machine learning (ML) techniques, which call for previous expert knowledge and expertise. This paper uses a deep learning (DL) framework based on 1D and 2D deep CNN with empirical mode decomposition (EMD) of a preprocessed ECG signal to propose a SA detection method to distinguish between a normal and apnea occurrence. The EMD is the perfect tool for removing crucial elements that characterise the underlying physiological or biological processes. Based on 5-fold cross-validation (5fold-CV), the segment-level classification performance had 93.8% accuracy with 94.9% sensitivity and 92.7% specificity. As a result, this work effectively created a unique and reliable SA detection system based on the ECG decomposed signal utilising EMD and deep CNN.
Introduction
I. INTRODUCTION
The prevalent sleep disease known as sleep apnea (SA) is characterised by frequent breathing pauses or bouts of shallow breathing during sleep that often go misdiagnosed and untreated [1]. During the course of an hour's worth of sleep at night, there may be five to over one hundred pauses, each lasting ten to twenty seconds. SA is associated with significant neurocognitive and cardiovascular consequences, and it may manifest with or without symptoms. Increased upper airway collapsibility during sleep causes considerably less airflow at the nose and/or mouth (hypopnea) or none at all (apnea). Oxyhemoglobin desaturation is generally present along with SA, which is usually interrupted by a short micro-arousal [2]. A protracted drop in oxyhemoglobin saturation and sleep fragmentation, with reduced slow-wave and rapid eye movement (REM) sleep, are the outcomes of recurrent apnea episodes.
SA is quite common, particularly in middle-aged and older adults. Based on the diagnostic criteria established by the American Academy of Sleep Apnea, 425 million individuals between the ages of 30 and 69 have moderate to severe SA (AASM) and about 936 million persons have mild to severe SA syndrome [3, 4].
Additional investigation into the correlation between the age and incidence of SA revealed that males 65 to 69 years old had an incidence of 88% events per hour or higher, and men 70 to 85 years old had an incidence of 90% events per hour. Furthermore, there is evidence connecting SA to a higher risk of several other illnesses, including heart failure, hypertension, heart attacks, and cardiovascular events.
Heart rate variability (HRV) and ECG waveform analysis have recently been established as an alternative technique to identify sleep disordered breathing, including apnea and hypopnea episodes [5,6]. Because the ECG signal is simple to collect with wearable technology and allows for the physiological evidence of SA incidence, it is a particularly intriguing signal to research. Blood oxygen levels fall during an apnea episode, and the cardiovascular system is activated to maintain the body's sufficient supply of oxygen. Therefore, elevated heart rate variability or aberrant cardiac activity may be signs of SA. The respiratory effort modifies the location of the ECG electrode, which affects the amplitude of the ECG signal in many ways [7]. In the meanwhile, the HRV evaluates the R-R interval, or variation in time between subsequent heartbeats (RRI). Since the fluctuation in the RRI is a sign of apnea episodes, it may provide the physiological foundation for identifying SA.
II. LITERATURE REVIEW
In this domain, machine learning classifiers such as Support Vector Machine (SVM) [8], k-Nearest Neighbours (kNN) [9], Random Forest (RF) [10], and deep learning classifiers like CNN are highly applicable, as OSA or any other type of apnea detection is a classification problem of the two classes—normal and apnea. The identification of sleep apnea has grown in importance as a study issue in the healthcare field, much like any other clinical diagnostic.
Using thoracic and abdominal signals as input characteristics, Ng et al. [11] have established a sensitivity of 70.29–86.25% for sleep apnea diagnosis. The non-linear analysis of blood oxygen saturation (Sa) derived from nocturnal oximetry has been studied by Alvarez et al. [12]. Based on the trials, 111 out of 187 participants tested positive for OSA. Qin et al.'s research [13] examined how OSA affected heart rate variability (HRV). They found that the degree of apnea illness tends to decrease HRV after conducting studies on 426 normal and 826 OSA afflicted people.
For the purpose of diagnosing apnea, several statistical body measurements, including as ECG, Sa, acoustic speech signal, and electroencephalogram (EEG), are available [14]. However, in our study, we have only concentrated on the ECG signal. Numerous studies on the diagnosis of apnea using ECG signals have previously been completed. Using the raw ECG signals from the PhysioNet Apnea-ECG database, Almazaydeh et al. [14] retrieved the relevant statistical characteristics, including mean, standard deviation, median, inter-quartile range, and part of their derivations for an RR interval (interval between two consecutive R peaks) [15]. With the help of SVM, they were able to extract these characteristics with an accuracy of up to 96.5%. Additionally, Cheng et al. [16] have experimented with the RR intervals of the PhysioNet Apnea-ECG database's ECG signal. Utilising the Recurrent Neural Network (RNN) [17], they were able to get findings that were 97.80% correct.
The Recurrence Quantification Analysis (RQA) statistics of the HRV data in the PhysioNet Apnea-ECG database were taken into consideration as characteristics by Nguyen et al. [18]. At first, they used both SVM and Artificial Neural Networks (ANN) to complete the classification job. They combined the scores from both classifiers using soft decision fusion, and the accuracy of the findings was 85.26%. The PhysioNet Apnea-ECG database's raw ECG signal was pre-processed by Hassan et al. [19] using the Tunable-Q factor Wavelet Transform (TQWT). They used an ensemble technique called Adaptive Boosting (AdaBoost) [20] for the decision tree, and 87.33% of the findings were correct.
Past time-windows have been taken into consideration by Wang et al. [21] for MLP architecture training. These time windows are limited to a minute in duration. Within each of these time windows, there are six time-domain RR Interval (RRI) features: mean of RRI (MRR), mean of heart rates (MHR), root mean square of differences between adjacent RRIs (RMSSD), standard deviation of RRIs (SDNN), number of adjacent RRIs exceeding 50 milliseconds (NN50), and pNN50 (NN50 divided by the number of RR intervals. Additionally, there are six frequency domain R-peak Amplitude features: Very Low Frequency (VLF), Low Frequency (LF), High Frequency (HF), LF/(LF + HF), and HF/(LF + HF). Finally, with an accuracy of 87.3%, they have produced the best outcome. In order to extract features from the RRI, Shen et al. [22] suggested MultiScale Dilation Attention 1-D CNN (MSDA-1DCNN). They also used a Weighted-Loss Time-Dependent (WLTD) classification model for OSA diagnosis, and using the PhysioNet Apnea-ECG database, they obtained 89.4% accuracy [15].
A unique 1-D CNN architecture has been presented by Chang et al. [23] for OSA detection. Each one-minute segment of the raw ECG data is first filtered using band pass, then Z-score normalisation is applied, and finally the CNN model is fitted. Overall, they have obtained an accuracy of 87.9% on the PhysioNet Apnea-ECG database [15], while for pre-recorded samples, the performance has climbed to 97.1%. A 1-D CNN architecture with a convolution layer, max pooling layer, fully linked MLP, and softmax output layer has been suggested by Thompson et al. [24]. Using window sizes of 500, 1000, 1500, 2000, and 2500, they used a windowing method in their study to validate their model, which on the PhysioNet Apnea-ECG database produced 93.77% accuracy for window size of 500 [15]. A unique CNN based on a scaleogram has been suggested by Mashrur et al. [25] to identify OSA from ECG data. Through the use of empirical mode decomposition (EMD) and continuous wavelet transform (CWT), they have successfully created hybrid scalograms from the ECG data. Using these scalograms, they train a CNN model to extract deep features for OSA detection, and using the PhysioNet Apnea-ECG database, they achieve an accuracy of 94.30% [15].
We chose to use ECG signal data for our study since most earlier efforts have taken the ECG into consideration. Another well-liked database for OSA detection research is PhysioNet Apnea-ECG. Since deep learning models are very suited to time-series data, we have selected them for our work [26]. Pre-processing is necessary since raw samples alone, as was mentioned in the Results and Discussion section, cannot fit CNN models to give exceptional results. Some of the known research based on ensemble methods are also covered, as our major focus is on the ensemble approaches in the apnea detection area.
On a given set of agents with their own nonlinear function approximator, such as an MLP, Faußer et al. [27] have applied Temporal Difference (TD) and Residual-Gradient (RG) update methods to adapt the weights to learn from joint decisions, such as Majority Voting and Averaging of the state-values. In order to develop a more stable and accurate model, Glodek et al. [28] have worked on ensemble techniques for density estimation utilising Gaussian Mixture Models (GMMs), which combine individual mixture models with a high degree of variability. In an ensemble of filter approaches, Chakraborty et al. [29] have built learning models utilising MLP-based classifiers with the chosen features, such as optimum subsets of features using filter methods Mutual Information (MI), Chi-square, and Anova F-Test.
Using an ensemble of RF and Radial Basis Function (RBF) networks, Kächele et al. [30] assessed the degree of discomfort in response to a video and physical characteristics like electromyography (EMG) and ECG. For score level fusion, they have trained using MLP using the classification results from each separate base model. To eliminate white Gaussian noise from a picture, Dey et al. [31] used a weighted ensemble of three CNN-based models: ADNet, IRCNN, and DnCNN. The outputs of the three models stated above are combined in the following ratios: 2:3:6. In order to quantify pain intensity levels efficiently, Bellmann et al. [32] have used a variety of fusion algorithms for the Multi-Classifier System (MCS). One of the most often used fusion techniques—bagging and boosting—is included in their case study. A fuzzy-rank based classifier fusion strategy has been presented by Kundu et al. [33].
The Gompartz function is used to determine the fuzzy-ranks of the basic classifiers. Using the ensemble of the pre-trained models VGG-11, Wide ResNet-50-2, and Inception v3, they were able to achieve the best results with 98.93% and 98.80% accuracies, respectively, in their experiments on the SARS-COV-2 [34] and Harvard Dataverse [35] datasets for diagnosing COVID-19 from the CT-scans.
The aforementioned established works demonstrate the broad range of study domains in which ensemble is used. The key factors contributing to classifier fusion's appeal are its enormous success and the breadth of its studies. As of right moment, no research using an ensemble approach has been done to diagnose apnea.This has inspired us to carry out ensemble-based experimental research in the OSA detection field. Furthermore, we have selected as base models the three deep learning models: (i) the CNN model offered by Wang et al. [36], (ii) the CNN model proposed by Sharan et al. [37], and (iii) the CNN-LSTM model proposed by Almutairi et al. [38]. This decision was made since all three of these models are CNN-based and, generally speaking, strong classifiers.
We are encouraged to work with the selected three models even more since they have been employed for OSA detection in the past. As a result, we used an ensemble of CNN-based designs in the widely used PhysioNet Apnea-ECG database to carry out our research [15].
III. METHODOLOGY
A. Overnight Apnea ECG Database
The performance of the suggested method was verified and made possible to be assessed and contrasted with the body of existing research using the Apnea ECG PhysioNet Database (AEPD), an accessible public database. Each participant's sleep recording lasted anywhere from seven to ten hours, with the AEPD ECG recordings captured at 100 Hz. Thirty-five subjects' minute-by-minute ECG recordings with SA comments were included in the database; each minute's recording was labelled as either normal breathing or disordered breathing with an episode of apnea.
The AEPD included three participant groups with 20, 5, and 10 participants each: A: apnea (Table 2), B: borderline (Table 3), and C: healthy control (Table 4). It was shown that members of Group A were connected to those who suffer from OSA, and each recording showed a cumulative apnea duration exceeding 100 minutes. Participants in this group varied in age from 38 to 63 and in AHI from 21 to 83.
The subjects in Group B were on the borderline, with total apnea episode lengths varying between 10 and 96 minutes. The age range of this group was 42–53, and the AHI was 0–25. Participants in Group C were healthy controls who had total apnea durations ranging from 0 to 3 minutes with either no SA or extremely low illness levels. Group C's recording of c05 and Group B's recording of b05 were removed because c05 was the same as c06 and b05 had a grinding noise.
After removing some contaminated events brought on by the patient's movement, inadequate patch contact, electrical interference, measurement noise, or other disturbances, only 33 recordings were included in this study, totaling 9160 normal events and 6019 apnea events (imbalanced dataset).
B. The Overall Deep Learning Framework for Automatic Sleep Apnea Classification Based on Empirical Mode Decomposition Derived from Single-Lead Electrocardiogram
The suggested sleep apnea detection system flowchart, which is based on the deep learning framework and the ECG signal decomposition, is shown in Figure 1. The automated SA classification system proposed in this paper is based on the deep learning framework and EMD, which is obtained from the single-lead ECG signal of both normal breathing and apnea. 33 individuals' overnight single-lead ECG signals were used as the input, along with each participant's 7–8 hour recordings, and were then divided into 60-second ECG windows. The members of AEPD groups A, B, and C were the participants in the mixed group category. Preprocessing steps for the raw ECG signal comprised windowing, band-pass filtering, and normalisation. To distinguish between SA and typical breathing episodes, decomposition characteristics were obtained using the EMD method. Next, a neighbourhood component analysis (NCA)-based feature selection method was used to minimise the amount of EMD features (IMFs). Four independent measures of fit (IMFs) were utilised in this study: IMF1, IMF2, IMF12 as a consequence of adding IMF1 and 2, and IMF123 as a result of adding IMF1, 2, and 3. Then, to address the issue of data imbalance, the synthetic minority oversampling method (SMOTE) was used. In the end, a 1D deep CNN model was created and cross-validated to get the classification results.
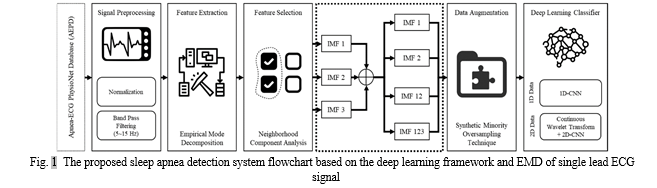
C. ECG Signal Preprocessing and Filtering
The ECG signal preprocessing included windowing processing settings and zero-means computing. In order to eliminate baseline wandering effects, this approach divides nocturnal ECG spectra into 60-s periods and excludes huge noise windows for algorithm development. Zero-means subtracts the mean from the ECG data. Each 60-second window was classified as either a normal episode or an apnea by the AEPD employed in this investigation.
IV. SYSTEM MODELLING
A. Overnight Apnea ECG Database
When creating a predictive model, feature selection is the process of minimising the number of input variables. Reducing the amount of input variables is preferable since it may lower modeling's computing cost and, in some situations, enhance the model's performance. A non-parametric technique called neighbourhood component analysis (NCA) is used to choose features in order to maximise the prediction accuracy of classification algorithms.
To get the best characteristics, the regularisation value (????) was adjusted. By eliminating unnecessary characteristics, NCA lowers the dimensionality of features and enhances algorithm performance. The following are the stages in NCA:
- Training and testing sets of data are separated apart. After that, the training data are divided into ten folds; the classifier is trained on nine of the folds and one is left out for testing.
- The NCA model was trained for each ???? using every fold in the training set, and the regularisation parameter, ???? value, is adjusted. This procedure is carried out once for every fold and every ???? value.
- Each fold's average loss was calculated for each ???? value, and the optimal ???? value—which is equivalent to the lowest average loss—was discovered.
- Lastly, characteristics that had feature weights higher than the threshold (T) were taken out.
????=????×max(????) in where ???? is the updated features weight and ???? is a tolerance set at 0.02.
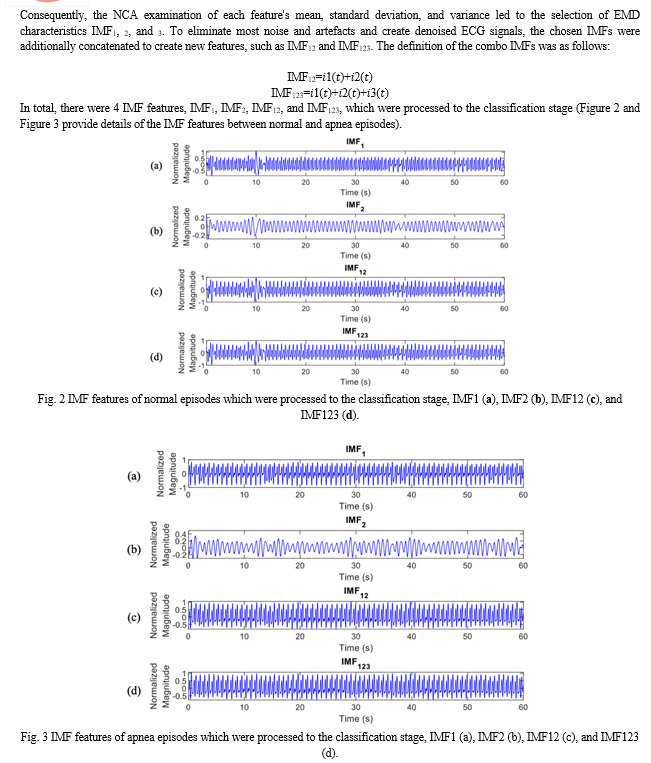
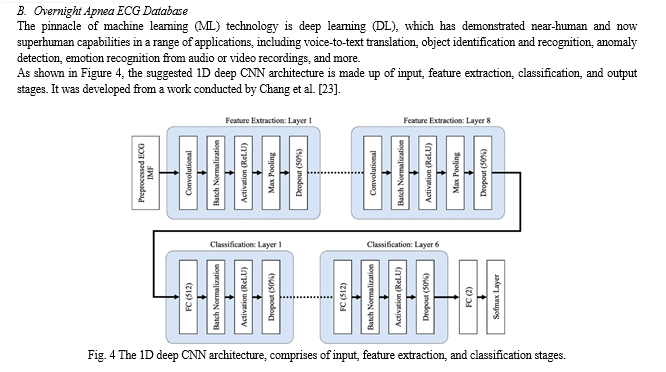
The CNN and FC layers both employ the He normal initialization approach to set the weights. By initialising the weights with consideration for the preceding layer of neurons, the cost function is able to approach the global minimum more quickly and effectively. The data is normalised before it reaches the ReLU activation layer by the batch normalisation layers, which are inserted after the CNN and FC layers in both the feature extraction and classification layers. This increases the neural network's speed, stability, and performance. The max pooling layers in the feature extraction layers minimise overfitting and network complexity by choosing the maximum activation regarding a neuron in a feature map. Each feature map's size is halved when the pooling size is set to 2. In order to minimise overfitting, 50% of the nodes are randomly excluded from dropout layers with a dropout rate of 0.5 during the training phase of the suggested CNN model. Testing accuracy is poor when overfitting is present, but training accuracy is high. Using the Adam optimizer, a stochastic gradient descent modification that calculates unique adaptive learning rates for various parameters based on estimations of the gradient's first and second moments, the suggested 1D deep CNN model was trained to minimise cross entropy.
C. Overnight Apnea ECG Database
Transfer learning is the process of using information from a previously learned network to improve learning in a new assignment. Transfer learning fine-tuning is usually more simpler and quicker than starting from scratch and training a network with randomly initialised weights. In this work, three different types of pretrained CNN models—AlexNet, GoogLeNet, and ResNet-50—were used for transfer learning. The key distinctions between AlexNet, GoogLeNet, and ResNet-50—the pretrained CNN models—are delineated in Table 1.
TABLE I
The main differences among pretrained 2D Deep CNN models that were used in this study
|
Characteristic Comparison |
2D Deep CNN Architecture |
||
|
AlexNet |
GoogLeNet |
ResNet-50 |
|
|
Number of Layers |
8 |
22 |
50 |
|
Input Size |
227×227×3 |
224×224×3 |
224×224×3 |
|
Number of Conv-Pool Layers |
5 |
21 |
49 |
|
Number of Fully Connected Layers |
3 |
1 |
1 |
|
Salient Feature |
Deeper |
Wider (parallel kernels) |
Shortcut connections |
|
Number of Parameters (Weights) |
60.97 million |
7 million |
25.56 million |
|
Memory |
232.5 MB |
26.3 MB |
97.2 MB |
|
FLOPs |
0.7×109 |
1.5×109 |
4.1×109 |
|
Training Time (5fold-CV/LOSO-CV) |
2.88/20.55 h |
4.56/25.41 h |
4.78/28.94 h |
Eight layers make up the AlexNet architecture: three fully connected layers, two ReLU layers, two Dropout layers for regularisation, a Softmax layer using a normalised exponential function, and three convolution layers (with two Convolution 2D layers, three Grouped Convolution 2D layers, five Rectified Linear Unit (ReLU) layers, two Cross-Channel Normalisation layers, and three Max-Pooling 2D layers). The creative part of the network lies in how effectively the ReLU activation function is implemented, together with how the Dropout mechanism and data augmentation approach are used to avoid overfitting. To improve model generalisation, the network has a Cross-Channel Normalisation layer. Moreover, the blurring effect caused by average pooling is removed by using maximum overlap pooling.
A pretrained CNN with 22 layers, including 9 inception levels, is called GoogLeNet. An inception layer in a convolutional vision network finds the best local sparse structure, which is then covered and approximated by easily accessible dense components. In order to avoid gradient disappearance, the network has an initial structure that increases the network's breadth and depth. The fully connected layer is removed and average pooling is used in its place.
A residual network (ResNet) constructed with 50 layers is referred to as ResNet-50. Presenting a "identity shortcut connection" that eschews one or more levels is the fundamental idea of a ResNet. By rerouting the input and expanding on the idea from the preceding layer, a shortcut connection, also known as a skip connection, is used to address the issue of disappearing or bursting gradients. During learning, a layer absorbs the ideas from the layer before it and combines with its inputs.
D. Cross-Validation
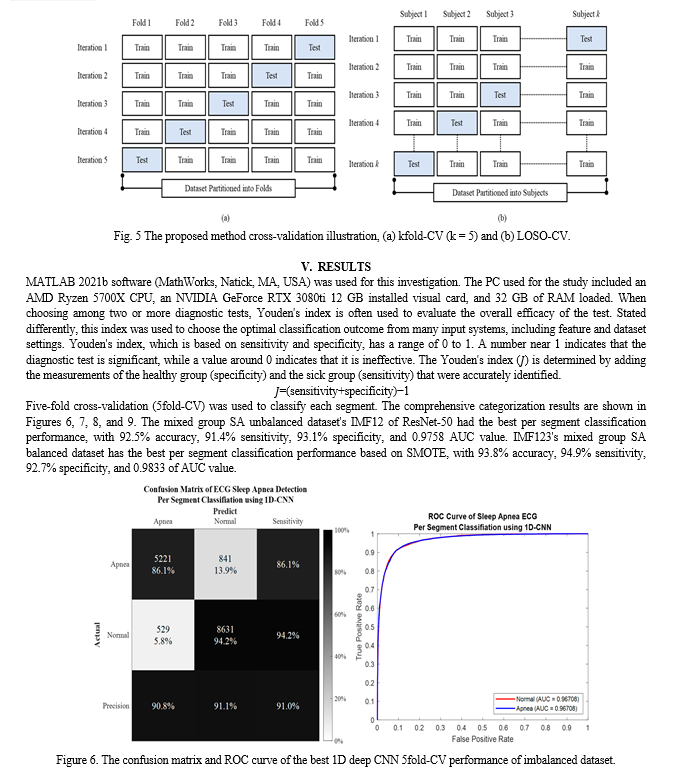
A statistical technique called cross-validation separates data into two groups: a training set (used for model training) and a testing set (used for model testing or validation). This allows for the evaluation and comparison of learning algorithms. To verify every data point, repeated rounds of cross-over between the training and testing sets are required. There are two primary uses for cross-validation: First, a single technique may be used to examine how well the learnt model performs with the data that is now available. Put otherwise, its purpose is to evaluate the generalizability of an algorithm. Finding the optimal algorithm for the provided data involves comparing the performance of two or more distinct algorithms, or, alternatively, comparing the performance of two or more parameterized model versions. Segment-level and subject-level validation were carried out using Leave-one-subject-out cross-validation (LOSO-CV) and k-fold cross-validation (kfold-CV), respectively.
The first step in the kfold-CV (Figure 5a) is to randomly divide the data samples into k subgroups. The remaining (k-1) subgroups may then be designated as the training set, and each subgroup can be selected as the testing set. This is how kfold-CV repeats testing and training k times; the average of the k accuracy scores for each iteration represents the final accuracy. In contrast, during every training procedure in the LOSO-CV, the data of a single subject are removed. Put differently, assume that there are N individuals in the dataset, that training is completed on N-1 subjects, and that validation is completed on a single subject. N times are spent repeating this procedure (Figure 5b).
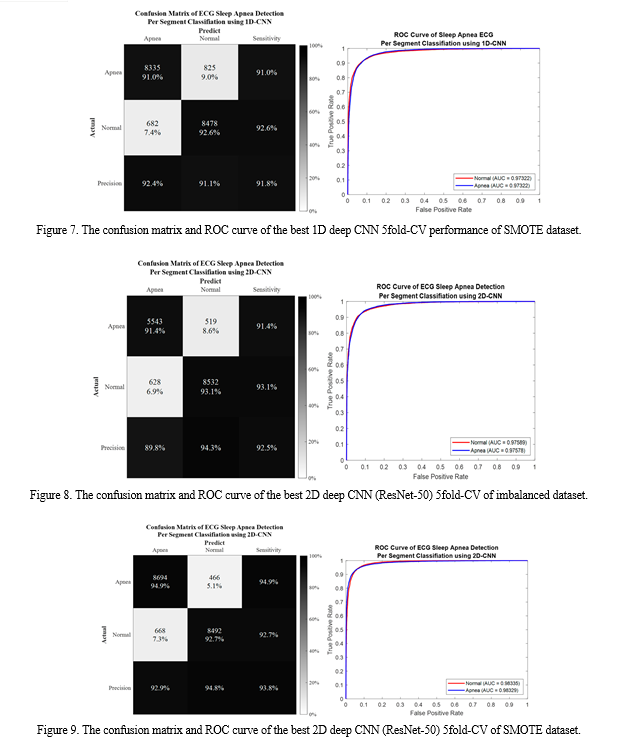
Conclusion
Using the signal decomposition approach, a unique algorithm was suggested in this article to categorise SA and normal breathing episodes. In addition to 1D and 2D deep learning classifiers, four distinct EMD characteristics were taken into account. High accuracy was achieved when the ECG signal of SA and typical breathing episodes was broken down. Furthermore, the algorithm performed more accurately than the state-of-the-art methods. In certain IMFs, the main characteristics of a normal and apnea episode are improved by using EMD to decompose the ECG signal. Furthermore, it was feasible to distinguish between normal and apnea episodes with success by converting the IMFs into the time-frequency spectrogram utilising CWT, pattern visualisation, and feature identification. Even though the suggested method worked very well, there are a few issues with this research. First, a small sample size from AEPD was used to verify the suggested technique. Second, since AEPD did not provide such information, inadequate physiological data and the subject\'s medical history were accessible for further investigation. These limitations may be addressed by combining several databases and gathering more information from the NCKUH Sleep Centre. Third, since the patients\' ECG signal was not only impacted by SA and was also somewhat erratic, the suggested method could not be used for patients experiencing the difficulties of cardiovascular disease (CVD). Future developments of this study include developing an algorithm to discriminate between SA and CVD using ECG data, testing the robustness of the proposed method in a large-scale group of participants from multiple databases, and automatically identifying physiological meanings for OSA features using explainable AI (XAI).
References
[1] Strollo, P.J., Jr.; Rogers, R.M. Obstructive sleep apnea. N. Engl. J. Med. 1996, 334, 99–104. [2] Memon, J.; Manganaro, S.N. Obstructive Sleep-disordered Breathing. In StatPearls [Internet]; StatPearls Publishing: Tampa, FL, USA, 2021. [3] Benjafield, A.V.; Ayas, N.T.; Eastwood, P.R.; Heinzer, R.; Ip, M.S.; Morrell, M.J.; Nunez, C.M.; Patel, S.R.; Penzel, T.; Pépin, J.-L. Estimation of the global prevalence and burden of obstructive sleep apnoea: A literature-based analysis. Lancet Respir. Med. 2019, 7, 687–698. [4] Kapur, V.K.; Auckley, D.H.; Chowdhuri, S.; Kuhlmann, D.C.; Mehra, R.; Ramar, K.; Harrod, C.G. Clinical practice guideline for diagnostic testing for adult obstructive sleep apnea: An American Academy of Sleep Medicine clinical practice guideline. J. Clin. Sleep Med. 2017, 13, 479–504. [5] Penzel, T.; Moody, G.B.; Mark, R.G.; Goldberger, A.L.; Peter, J.H. The apnea-ECG database. Proc. Comput. Cardiol. 2000, 27, 255–258. [6] Penzel, T.; Kantelhardt, J.W.; Grote, L.; Peter, J.-H.; Bunde, A. Comparison of detrended fluctuation analysis and spectral analysis for heart rate variability in sleep and sleep apnea. IEEE Trans. Biomed. Eng. 2003, 50, 1143–1151. [7] Janbakhshi, P.; Shamsollahi, M.B. ECG-derived respiration estimation from single-lead ECG using gaussian process and phase space reconstruction methods. Biomed. Signal Processing Control 2018, 45, 80–90. [8] Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [9] Guo, G.; Wang, H.; Bell, D.; Bi, Y.; Greer, K. KNN model-based approach in classification. In Lecture Notes in Computer Science, Proceedings of the OTM Confederated International Conferences “On the Move to Meaningful Internet Systems”, Catania, Italy, 3–7 November 2003; Springer: Berlin/Heidelberg, Germany, 2003; pp. 986–996. [10] Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [11] Ng, A.S.; Chung, J.W.; Gohel, M.D.; Yu, W.W.; Fan, K.L.; Wong, T.K. Evaluation of the performance of using mean absolute amplitude analysis of thoracic and abdominal signals for immediate indication of sleep apnoea events. J. Clin. Nurs. 2008, 17, 2360–2366. [12] Alvarez, D.; Hornero, R.; Abásolo, D.; Del Campo, F.; Zamarrón, C. Nonlinear characteristics of blood oxygen saturation from nocturnal oximetry for obstructive sleep apnoea detection. Physiol. Meas. 2006, 27, 399. [13] Qin, H.; Keenan, B.T.; Mazzotti, D.R.; Vaquerizo-Villar, F.; Kraemer, J.F.; Wessel, N.; Tufik, S.; Bittencourt, L.; Cistulli, P.A.; de Chazal, P.; et al. Heart rate variability during wakefulness as a marker of obstructive sleep apnea severity. Sleep 2021, 44, zsab018. [14] Almazaydeh, L.; Elleithy, K.; Faezipour, M. Detection of obstructive sleep apnea through ECG signal features. In Proceedings of the 2012 IEEE International Conference on Electro/Information Technology, Indianapolis, IN, USA, 6–8 May 2012; pp. 1–6. [15] Penzel, T.; Moody, G.B.; Mark, R.G.; Goldberger, A.L.; Peter, J.H. The apnea-ECG database. In Proceedings of the Computers in Cardiology 2000. Vol. 27 (Cat. 00CH37163), Cambridge, MA, USA, 24–27 September 2000; pp. 255–258. [16] Cheng, M.; Sori, W.J.; Jiang, F.; Khan, A.; Liu, S. Recurrent neural network based classification of ECG signal features for obstruction of sleep apnea detection. In Proceedings of the 2017 IEEE International Conference on Computational Science and Engineering (CSE) and IEEE International Conference on Embedded and Ubiquitous Computing (EUC), Guangzhou, China, 21–24 July 2017; Volume 2, pp. 199–202. [17] Marhon, S.A.; Cameron, C.J.; Kremer, S.C. Recurrent neural networks. In Handbook on Neural Information Processing; Springer: Berlin/Heidelberg, Germany, 2013; pp. 29–65. [18] Nguyen, H.D.; Wilkins, B.A.; Cheng, Q.; Benjamin, B.A. An online sleep apnea detection method based on recurrence quantification analysis. IEEE J. Biomed. Health Inform. 2013, 18, 1285–1293. [19] Hassan, A.R. Computer-aided obstructive sleep apnea detection using normal inverse Gaussian parameters and adaptive boosting. Biomed. Signal Process. Control 2016, 29, 22–30. [20] Freund, Y.; Schapire, R.E. A decision-theoretic generalization of on-line learning and an application to boosting. J. Comput. Syst. Sci. 1997, 55, 119–139. [21] Wang, T.; Lu, C.; Shen, G. Detection of sleep apnea from single-lead ECG signal using a time window artificial neural network. BioMed Res. Int. 2019, 2019. [22] Shen, Q.; Qin, H.; Wei, K.; Liu, G. Multiscale Deep Neural Network for Obstructive Sleep Apnea Detection Using RR Interval From Single-Lead ECG Signal. IEEE Trans. Instrum. Meas. 2021, 70, 1–13. [23] Chang, H.Y.; Yeh, C.Y.; Lee, C.T.; Lin, C.C. A sleep apnea detection system based on a one-dimensional deep convolution neural network model using single-lead electrocardiogram. Sensors 2020, 20, 4157. [24] Thompson, S.; Fergus, P.; Chalmers, C.; Reilly, D. Detection of Obstructive Sleep Apnoea Using Features Extracted from Segmented Time-Series ECG Signals Using a One Dimensional Convolutional Neural Network. In Proceedings of the 2020 International Joint Conference on Neural Networks (IJCNN), Glasgow, UK, 19–24 July 2020; pp. 1–8. [25] Mashrur, F.R.; Islam, M.S.; Saha, D.K.; Islam, S.R.; Moni, M.A. SCNN: Scalogram-based Convolutional Neural Network to Detect Obstructive Sleep Apnea using Single-lead Electrocardiogram Signals. Comput. Biol. Med. 2021, 134, 104532. [26] Zhao, B.; Lu, H.; Chen, S.; Liu, J.; Wu, D. Convolutional neural networks for time series classification. J. Syst. Eng. Electron. 2017, 28, 162–169. [27] Faußer, S.; Schwenker, F. Ensemble methods for reinforcement learning with function approximation. In Lecture Notes in Computer Science, Proceedings of the International Workshop on Multiple Classifier Systems, Naples, Italy, 15–17 June 2011; Springer: Berlin/Heidelberg, Germany, 2011; pp. 56–65. [28] Glodek, M.; Schels, M.; Schwenker, F. Ensemble Gaussian mixture models for probability density estimation. Comput. Stat. 2013, 28, 127–138. [29] Chakraborty, A.; De, R.; Chatterjee, A.; Schwenker, F.; Sarkar, R. Filter Method Ensemble with Neural Networks. In Lecture Notes in Computer Science, Proceedings of the International Conference on Artificial Neural Networks, Munich, Germany, 17–19 September 2019; Springer: Cham, Switzerland, 2019; pp. 755–765. [30] Kächele, M.; Amirian, M.; Thiam, P.; Werner, P.; Walter, S.; Palm, G.; Schwenker, F. Adaptive confidence learning for the personalization of pain intensity estimation systems. Evol. Syst. 2017, 8, 71–83. [31] Dey, S.; Bhattacharya, R.; Schwenker, F.; Sarkar, R. Median Filter Aided CNN Based Image Denoising: An Ensemble Aprroach. Algorithms 2021, 14, 109. [32] Bellmann, P.; Thiam, P.; Schwenker, F. Multi-classifier-systems: Architectures, algorithms and applications. In Computational Intelligence for Pattern Recognition; Springer: Berlin/Heidelberg, Germany, 2018; pp. 83–113. [33] Kundu, R.; Basak, H.; Singh, P.K.; Ahmadian, A.; Ferrara, M.; Sarkar, R. Fuzzy rank-based fusion of CNN models using Gompertz function for screening COVID-19 CT-scans. Sci. Rep. 2021, 11, 1–12. [34] Angelov, P.; Almeida Soares, E. SARS-CoV-2 CT-scan dataset: A large dataset of real patients CT scans for SARS-CoV-2 identification. MedRxiv 2020. [35] Soares, E.; Angelov, P. A large dataset of real patients CT scans for COVID-19 identification. Harv. Dataverse 2020, 1. [36] Wang, X.; Cheng, M.; Wang, Y.; Liu, S.; Tian, Z.; Jiang, F.; Zhang, H. Obstructive sleep apnea detection using ecg-sensor with convolutional neural networks. Multimed. Tools Appl. 2020, 79, 15813–15827. [37] Sharan, R.V.; Berkovsky, S.; Xiong, H.; Coiera, E. ECG-Derived Heart Rate Variability Interpolation and 1-D Convolutional Neural Networks for Detecting Sleep Apnea. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 637–640. [38] Almutairi, H.; Hassan, G.M.; Datta, A. Detection of Obstructive Sleep Apnoea by ECG signals using Deep Learning Architectures. In Proceedings of the 2020 28th European Signal Processing Conference (EUSIPCO), Amsterdam, The Netherlands, 18–21 January 2021; pp. 1382–1386.
Copyright
Copyright © 2024 R. Krithiga Sree, Dr. D. Somasundareswari. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET63676
Publish Date : 2024-07-19
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

